State of DNA Storage Discussed in New Whitepaper

(CI Photos/Shutterstock)
The DNA Storage Alliance, which is part of the SNIA community, last month released a technical whitepaper detailing the progress so far in creating DNA storage, when we might see commercial DNA storage offerings, and the technical hurdles that remain.
Thanks to its incredible density, universal applicability, and multi-century lifespan, DNA storage is viewed by some as a possible storage medium for the future. The idea is gaining new backing as an archive of last resort, particularly as the AI revolution surges into high gear, leading people find greater value in data that they previously would have discarded.
The current data dynamic is putting pressure on traditional storage technologies, which aren’t keeping up with surging data retention rates, the DNA Storage Alliance says in its new whitepaper, titled “DNA Data Storage Technology Review.”
“The capital costs associated with traditional storage media are not scaling with the rate of data generation, and operational costs of refreshing data, or creating copies, using existing storage technologies is becoming prohibitive,” the whitepaper authors write. “The rate of HDD and tape media storage density growth is slowing, and media lifetime is not improving significantly.”
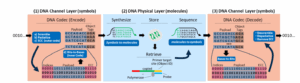
DNA storage provides one alternative storage method that could meet the data storage tsunami. Here’s how it works at a high level: Binary data is encoded into ACGT language of DNA–(A), cytosine (C), guanine (G), and thymine (T)–where it is translated into molecular strings stored in DNA. To retrieve the stored data, the process is reversed.
While the general “bits to bases” and “bases to bits” theory holds true, there are certain limitations to keep in mind with this process, the DNA Storage Alliance writes in its paper. For starters, there is a need to correct for various physical errors that may occur in the DNA “physical layer.” Certain sequences should also be avoided due to the probability of causing synthesis or sequencing errors.
There are also various ways that practitioners can encode the DNA data. The most basic path is a straight binary representation, where A, C, G, and T are encoded as 00, 01, 10, and 11. There is also the ternary representation, which uses a base-3 encoding, which avoids generating strings of repeated bases, or homopolymers. Practitioners can also use combinatorial assembly that uses short DNA sequences as building blocks to create bigger DNA molecules, which can provide a full alphabet of data encoders.
Another technique is called topological modification, where the data is converted into a positional structure, which the authors describe as a “DNA punch card” (but please don’t tell the mainframers). DNA nanostructures can also be used to encode data, and there is also the potential to create composite DNA letters, which further increases the size of the available alphabet.
Like all forms of storage, DNA storage requires error correction. DNA synthesis has about a 1% natural error rate, but there are various approaches to correct them, including use of parity codes, low density parity checks, CRC checksums, erasure codes, fountain codes, and Viterbi codes. Depending on the situation, a practitioner might combine multiple of these error correction codes together into a single process, according to the whitepaper authors.
Some DNA patterns can increase the odds of an error, which is why DNA storage must incorporate constraints to avoid them. The whitepaper authors discuss various strategies for building these constraints, including local and global constraints, as well as biosafety and biosecurity concerns.
Other aspects to consider for DNA storage are the various data storage protocols that need to be built and implemented. For instance, since a single GB may not fit into a DNA molecule, the source data object must be broken up into smaller pieces, or packetized. DNA is universal, which is one of its big selling points, but certain care must be taken to ensure that the data can be explored in a DNA archive, the authors write. Finally, tags will be used in DNA storage to speed up random file access.
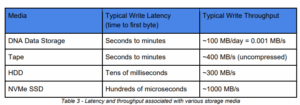
Like other storage mediums, DNA storage will function at a certain throughput and with certain latencies. On the latency front, current DNA storage writes to media at about the same rate as it does to tape, in seconds to minutes. On the throughput front, DNA storage will be able to handle about 100 MB per day, which is orders of magnitude slower than tape (about 400 MB per second), let alone NVMe disk.
Speed obviously won’t be DNA storage’s strong suit, at least as it’s currently envisioned. However, when kept away from sunlight, water, and air, DNA storage has the potential to store data for a very long time. The whitepaper authors remind us that DNA has been recovered from fossilized mammals that were 2 million years old. Most organizations would probably be happy with a couple of decades.
To preserve DNA, one must create a DNA Containment System (DCS), which utilizes vessels equipped with seals, additives, and inert gases, the authors tell us. Currently, the cost of implementing error correction is limiting DNA storage to single strands that store tens of bytes. Storing bigger data sets will require encoding into DNA sub-strands and using encoding indices, the authors write.
Within three to five years, DNA storage will be viable for archival use cases, the DNA Storage Alliance authors write.
“It is important to see DNA data storage not as a replacement for any existing storage technology, but as a complementary capability that enables the data storage hierarchy to expand, resolving the ‘save/discard’ dilemma with a viable TCO for zettabyte scale and data preservation,” they write. “While DNA data storage is still quite nascent and there remain significant challenges to commercialization, the foundations of writing, storing, retrieving, and reading data using DNA have been shown to work on scalable technology platforms.”
The DNA Storage Alliance, which is a component of the Storage Networking Industry Association (SNIA), has dozens of members, including Western Digital, Twist Bioscience, Catalog, Imagene, Biomemory, Los Alamos National Laboratory, Kioxia, Dell Technologies, Seagate, IBM, and others.
You can download the whitepaper here.
Related Items:
Harvard’s New Data Storage Is to Dye For, Avoids DNA Storage Pitfalls
Storage Approach Mimics DNA in Fossils